Build regression model from a set of candidate predictor variables by entering predictors based on p values, in a stepwise manner until there is no variable left to enter any more.
Usage
ols_step_forward_p(model, ...)
# Default S3 method
ols_step_forward_p(
model,
p_val = 0.3,
include = NULL,
exclude = NULL,
hierarchical = FALSE,
progress = FALSE,
details = FALSE,
...
)
# S3 method for class 'ols_step_forward_p'
plot(x, model = NA, print_plot = TRUE, details = TRUE, ...)Arguments
- model
An object of class
lm; the model should include all candidate predictor variables.- ...
Other arguments.
- p_val
p value; variables with p value less than
p_valwill enter into the model- include
Character or numeric vector; variables to be included in selection process.
- exclude
Character or numeric vector; variables to be excluded from selection process.
- hierarchical
Logical; if
TRUE, performs hierarchical selection.- progress
Logical; if
TRUE, will display variable selection progress.- details
Logical; if
TRUE, will print the regression result at each step.- x
An object of class
ols_step_forward_p.- print_plot
logical; if
TRUE, prints the plot else returns a plot object.
Value
ols_step_forward_p returns an object of class "ols_step_forward_p".
An object of class "ols_step_forward_p" is a list containing the
following components:
- model
final model; an object of class
lm- metrics
selection metrics
References
Chatterjee, Samprit and Hadi, Ali. Regression Analysis by Example. 5th ed. N.p.: John Wiley & Sons, 2012. Print.
Kutner, MH, Nachtscheim CJ, Neter J and Li W., 2004, Applied Linear Statistical Models (5th edition). Chicago, IL., McGraw Hill/Irwin.
See also
Other forward selection procedures:
ols_step_forward_adj_r2(),
ols_step_forward_aic(),
ols_step_forward_r2(),
ols_step_forward_sbc(),
ols_step_forward_sbic()
Examples
# stepwise forward regression
model <- lm(y ~ ., data = surgical)
ols_step_forward_p(model)
#>
#>
#> Stepwise Summary
#> --------------------------------------------------------------------------
#> Step Variable AIC SBC SBIC R2 Adj. R2
#> --------------------------------------------------------------------------
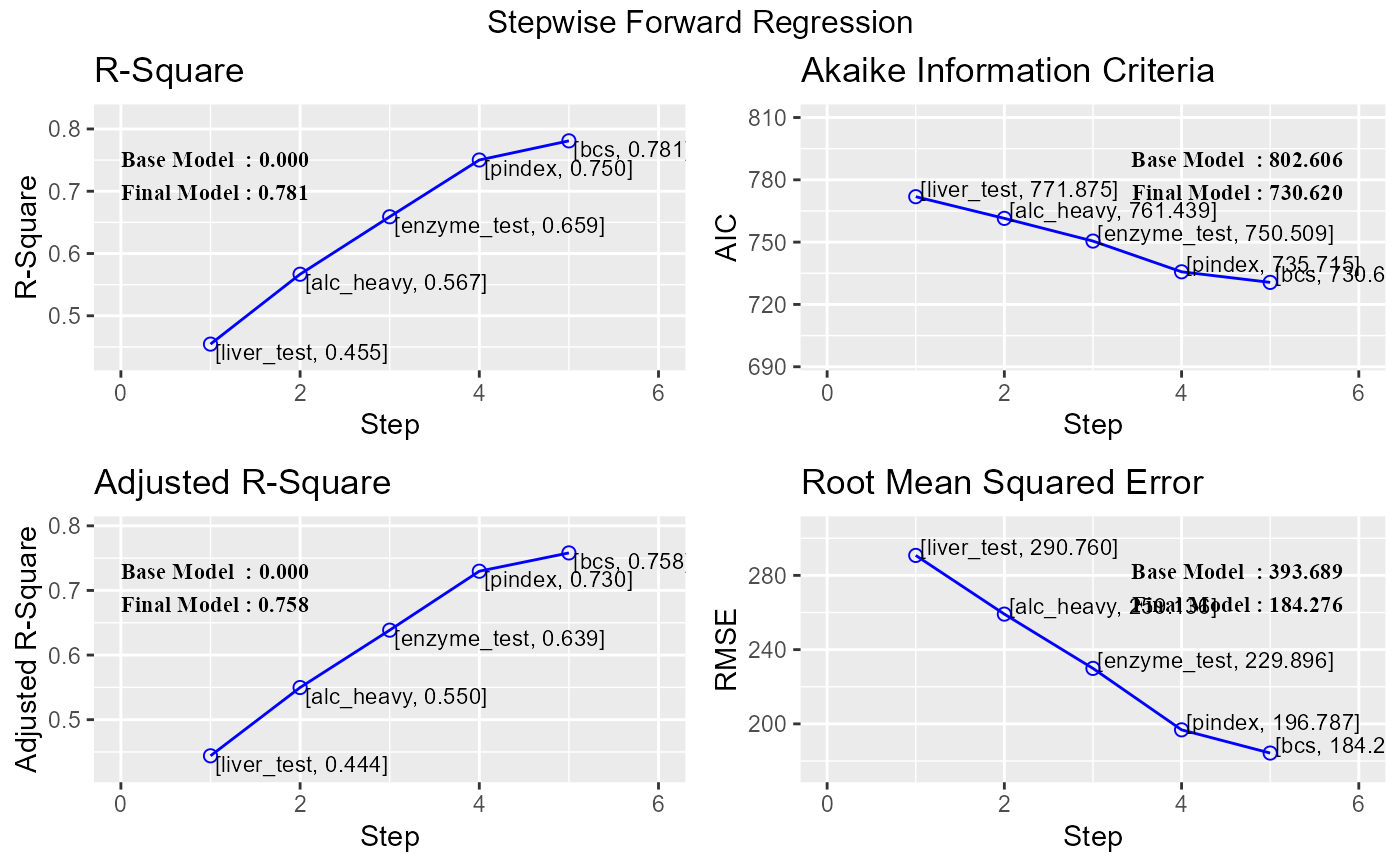
#> 0 Base Model 802.606 806.584 646.794 0.00000 0.00000
#> 1 liver_test 771.875 777.842 616.009 0.45454 0.44405
#> 2 alc_heavy 761.439 769.395 605.506 0.56674 0.54975
#> 3 enzyme_test 750.509 760.454 595.297 0.65900 0.63854
#> 4 pindex 735.715 747.649 582.943 0.75015 0.72975
#> 5 bcs 730.620 744.543 579.638 0.78091 0.75808
#> --------------------------------------------------------------------------
#>
#> Final Model Output
#> ------------------
#>
#> Model Summary
#> -------------------------------------------------------------------
#> R 0.884 RMSE 184.276
#> R-Squared 0.781 MSE 33957.712
#> Adj. R-Squared 0.758 Coef. Var 27.839
#> Pred R-Squared 0.700 AIC 730.620
#> MAE 137.656 SBC 744.543
#> -------------------------------------------------------------------
#> RMSE: Root Mean Square Error
#> MSE: Mean Square Error
#> MAE: Mean Absolute Error
#> AIC: Akaike Information Criteria
#> SBC: Schwarz Bayesian Criteria
#>
#> ANOVA
#> -----------------------------------------------------------------------
#> Sum of
#> Squares DF Mean Square F Sig.
#> -----------------------------------------------------------------------
#> Regression 6535804.090 5 1307160.818 34.217 0.0000
#> Residual 1833716.447 48 38202.426
#> Total 8369520.537 53
#> -----------------------------------------------------------------------
#>
#> Parameter Estimates
#> ------------------------------------------------------------------------------------------------
#> model Beta Std. Error Std. Beta t Sig lower upper
#> ------------------------------------------------------------------------------------------------
#> (Intercept) -1178.330 208.682 -5.647 0.000 -1597.914 -758.746
#> liver_test 58.064 40.144 0.156 1.446 0.155 -22.652 138.779
#> alc_heavy 317.848 71.634 0.314 4.437 0.000 173.818 461.878
#> enzyme_test 9.748 1.656 0.521 5.887 0.000 6.419 13.077
#> pindex 8.924 1.808 0.380 4.935 0.000 5.288 12.559
#> bcs 59.864 23.060 0.241 2.596 0.012 13.498 106.230
#> ------------------------------------------------------------------------------------------------
#>
# stepwise forward regression plot
model <- lm(y ~ ., data = surgical)
k <- ols_step_forward_p(model)
plot(k)
 # selection metrics
k$metrics
#> step variable r2 adj_r2 aic sbc sbic mallows_cp
#> 1 1 liver_test 0.4545389 0.4440492 771.8753 777.8423 616.0089 62.511923
#> 2 2 alc_heavy 0.5667409 0.5497504 761.4394 769.3953 605.5062 41.368078
#> 3 3 enzyme_test 0.6590000 0.6385400 750.5089 760.4538 595.2974 24.337853
#> 4 4 pindex 0.7501457 0.7297495 735.7146 747.6485 582.9426 7.537284
#> 5 5 bcs 0.7809054 0.7580831 730.6204 744.5433 579.6377 3.192498
#> rmse
#> 1 290.7604
#> 2 259.1357
#> 3 229.8956
#> 4 196.7872
#> 5 184.2762
# final model
k$model
#>
#> Call:
#> lm(formula = paste(response, "~", paste(preds, collapse = " + ")),
#> data = l)
#>
#> Coefficients:
#> (Intercept) liver_test alc_heavy enzyme_test pindex bcs
#> -1178.330 58.064 317.848 9.748 8.924 59.864
#>
# include or exclude variables
# force variable to be included in selection process
ols_step_forward_p(model, include = c("age", "alc_mod"))
#>
#>
#> Stepwise Summary
#> ---------------------------------------------------------------------------
#> Step Variable AIC SBC SBIC R2 Adj. R2
#> ---------------------------------------------------------------------------
#> 0 Base Model 804.340 812.295 645.675 0.04110 0.00350
#> 1 age 803.834 809.801 646.572 0.01420 -0.00476
#> 2 alc_mod 804.340 812.295 645.675 0.04110 0.00350
#> 3 liver_test 772.922 782.867 615.246 0.48357 0.45258
#> 4 enzyme_test 763.665 775.599 606.382 0.58074 0.54652
#> 5 alc_heavy 754.332 768.255 598.224 0.66012 0.62471
#> 6 pindex 739.680 755.592 587.108 0.75031 0.71843
#> ---------------------------------------------------------------------------
#>
#> Final Model Output
#> ------------------
#>
#> Model Summary
#> -------------------------------------------------------------------
#> R 0.866 RMSE 196.724
#> R-Squared 0.750 MSE 38700.429
#> Adj. R-Squared 0.718 Coef. Var 30.034
#> Pred R-Squared 0.649 AIC 739.680
#> MAE 146.418 SBC 755.592
#> -------------------------------------------------------------------
#> RMSE: Root Mean Square Error
#> MSE: Mean Square Error
#> MAE: Mean Absolute Error
#> AIC: Akaike Information Criteria
#> SBC: Schwarz Bayesian Criteria
#>
#> ANOVA
#> -----------------------------------------------------------------------
#> Sum of
#> Squares DF Mean Square F Sig.
#> -----------------------------------------------------------------------
#> Regression 6279697.346 6 1046616.224 23.538 0.0000
#> Residual 2089823.191 47 44464.323
#> Total 8369520.537 53
#> -----------------------------------------------------------------------
#>
#> Parameter Estimates
#> -----------------------------------------------------------------------------------------------
#> model Beta Std. Error Std. Beta t Sig lower upper
#> -----------------------------------------------------------------------------------------------
#> (Intercept) -814.092 213.222 -3.818 0.000 -1243.041 -385.144
#> age 0.458 2.706 0.013 0.169 0.866 -4.985 5.902
#> alc_mod 1.088 67.941 0.001 0.016 0.987 -135.591 137.768
#> liver_test 126.675 33.832 0.341 3.744 0.000 58.613 194.737
#> enzyme_test 7.523 1.543 0.402 4.874 0.000 4.418 10.628
#> alc_heavy 361.751 87.140 0.357 4.151 0.000 186.448 537.053
#> pindex 7.862 1.908 0.334 4.120 0.000 4.023 11.700
#> -----------------------------------------------------------------------------------------------
#>
# use index of variable instead of name
ols_step_forward_p(model, include = c(5, 7))
#>
#>
#> Stepwise Summary
#> ---------------------------------------------------------------------------
#> Step Variable AIC SBC SBIC R2 Adj. R2
#> ---------------------------------------------------------------------------
#> 0 Base Model 804.340 812.295 645.675 0.04110 0.00350
#> 1 age 803.834 809.801 646.572 0.01420 -0.00476
#> 2 alc_mod 804.340 812.295 645.675 0.04110 0.00350
#> 3 liver_test 772.922 782.867 615.246 0.48357 0.45258
#> 4 enzyme_test 763.665 775.599 606.382 0.58074 0.54652
#> 5 alc_heavy 754.332 768.255 598.224 0.66012 0.62471
#> 6 pindex 739.680 755.592 587.108 0.75031 0.71843
#> ---------------------------------------------------------------------------
#>
#> Final Model Output
#> ------------------
#>
#> Model Summary
#> -------------------------------------------------------------------
#> R 0.866 RMSE 196.724
#> R-Squared 0.750 MSE 38700.429
#> Adj. R-Squared 0.718 Coef. Var 30.034
#> Pred R-Squared 0.649 AIC 739.680
#> MAE 146.418 SBC 755.592
#> -------------------------------------------------------------------
#> RMSE: Root Mean Square Error
#> MSE: Mean Square Error
#> MAE: Mean Absolute Error
#> AIC: Akaike Information Criteria
#> SBC: Schwarz Bayesian Criteria
#>
#> ANOVA
#> -----------------------------------------------------------------------
#> Sum of
#> Squares DF Mean Square F Sig.
#> -----------------------------------------------------------------------
#> Regression 6279697.346 6 1046616.224 23.538 0.0000
#> Residual 2089823.191 47 44464.323
#> Total 8369520.537 53
#> -----------------------------------------------------------------------
#>
#> Parameter Estimates
#> -----------------------------------------------------------------------------------------------
#> model Beta Std. Error Std. Beta t Sig lower upper
#> -----------------------------------------------------------------------------------------------
#> (Intercept) -814.092 213.222 -3.818 0.000 -1243.041 -385.144
#> age 0.458 2.706 0.013 0.169 0.866 -4.985 5.902
#> alc_mod 1.088 67.941 0.001 0.016 0.987 -135.591 137.768
#> liver_test 126.675 33.832 0.341 3.744 0.000 58.613 194.737
#> enzyme_test 7.523 1.543 0.402 4.874 0.000 4.418 10.628
#> alc_heavy 361.751 87.140 0.357 4.151 0.000 186.448 537.053
#> pindex 7.862 1.908 0.334 4.120 0.000 4.023 11.700
#> -----------------------------------------------------------------------------------------------
#>
# force variable to be excluded from selection process
ols_step_forward_p(model, exclude = c("pindex"))
#>
#>
#> Stepwise Summary
#> --------------------------------------------------------------------------
#> Step Variable AIC SBC SBIC R2 Adj. R2
#> --------------------------------------------------------------------------
#> 0 Base Model 802.606 806.584 646.794 0.00000 0.00000
#> 1 liver_test 771.875 777.842 616.009 0.45454 0.44405
#> 2 alc_heavy 761.439 769.395 605.506 0.56674 0.54975
#> 3 enzyme_test 750.509 760.454 595.297 0.65900 0.63854
#> 4 bcs 750.782 762.716 595.377 0.66973 0.64277
#> --------------------------------------------------------------------------
#>
#> Final Model Output
#> ------------------
#>
#> Model Summary
#> -------------------------------------------------------------------
#> R 0.818 RMSE 226.248
#> R-Squared 0.670 MSE 51188.335
#> Adj. R-Squared 0.643 Coef. Var 33.829
#> Pred R-Squared 0.567 AIC 750.782
#> MAE 171.544 SBC 762.716
#> -------------------------------------------------------------------
#> RMSE: Root Mean Square Error
#> MSE: Mean Square Error
#> MAE: Mean Absolute Error
#> AIC: Akaike Information Criteria
#> SBC: Schwarz Bayesian Criteria
#>
#> ANOVA
#> -----------------------------------------------------------------------
#> Sum of
#> Squares DF Mean Square F Sig.
#> -----------------------------------------------------------------------
#> Regression 5605350.444 4 1401337.611 24.841 0.0000
#> Residual 2764170.093 49 56411.635
#> Total 8369520.537 53
#> -----------------------------------------------------------------------
#>
#> Parameter Estimates
#> ----------------------------------------------------------------------------------------------
#> model Beta Std. Error Std. Beta t Sig lower upper
#> ----------------------------------------------------------------------------------------------
#> (Intercept) -534.711 197.967 -2.701 0.009 -932.540 -136.882
#> liver_test 149.496 43.277 0.403 3.454 0.001 62.528 236.464
#> alc_heavy 292.382 86.822 0.288 3.368 0.001 117.907 466.858
#> enzyme_test 7.431 1.929 0.397 3.851 0.000 3.554 11.309
#> bcs 34.471 27.316 0.139 1.262 0.213 -20.422 89.365
#> ----------------------------------------------------------------------------------------------
#>
# use index of variable instead of name
ols_step_forward_p(model, exclude = c(2))
#>
#>
#> Stepwise Summary
#> --------------------------------------------------------------------------
#> Step Variable AIC SBC SBIC R2 Adj. R2
#> --------------------------------------------------------------------------
#> 0 Base Model 802.606 806.584 646.794 0.00000 0.00000
#> 1 liver_test 771.875 777.842 616.009 0.45454 0.44405
#> 2 alc_heavy 761.439 769.395 605.506 0.56674 0.54975
#> 3 enzyme_test 750.509 760.454 595.297 0.65900 0.63854
#> 4 bcs 750.782 762.716 595.377 0.66973 0.64277
#> --------------------------------------------------------------------------
#>
#> Final Model Output
#> ------------------
#>
#> Model Summary
#> -------------------------------------------------------------------
#> R 0.818 RMSE 226.248
#> R-Squared 0.670 MSE 51188.335
#> Adj. R-Squared 0.643 Coef. Var 33.829
#> Pred R-Squared 0.567 AIC 750.782
#> MAE 171.544 SBC 762.716
#> -------------------------------------------------------------------
#> RMSE: Root Mean Square Error
#> MSE: Mean Square Error
#> MAE: Mean Absolute Error
#> AIC: Akaike Information Criteria
#> SBC: Schwarz Bayesian Criteria
#>
#> ANOVA
#> -----------------------------------------------------------------------
#> Sum of
#> Squares DF Mean Square F Sig.
#> -----------------------------------------------------------------------
#> Regression 5605350.444 4 1401337.611 24.841 0.0000
#> Residual 2764170.093 49 56411.635
#> Total 8369520.537 53
#> -----------------------------------------------------------------------
#>
#> Parameter Estimates
#> ----------------------------------------------------------------------------------------------
#> model Beta Std. Error Std. Beta t Sig lower upper
#> ----------------------------------------------------------------------------------------------
#> (Intercept) -534.711 197.967 -2.701 0.009 -932.540 -136.882
#> liver_test 149.496 43.277 0.403 3.454 0.001 62.528 236.464
#> alc_heavy 292.382 86.822 0.288 3.368 0.001 117.907 466.858
#> enzyme_test 7.431 1.929 0.397 3.851 0.000 3.554 11.309
#> bcs 34.471 27.316 0.139 1.262 0.213 -20.422 89.365
#> ----------------------------------------------------------------------------------------------
#>
# hierarchical selection
model <- lm(y ~ bcs + alc_heavy + pindex + enzyme_test, data = surgical)
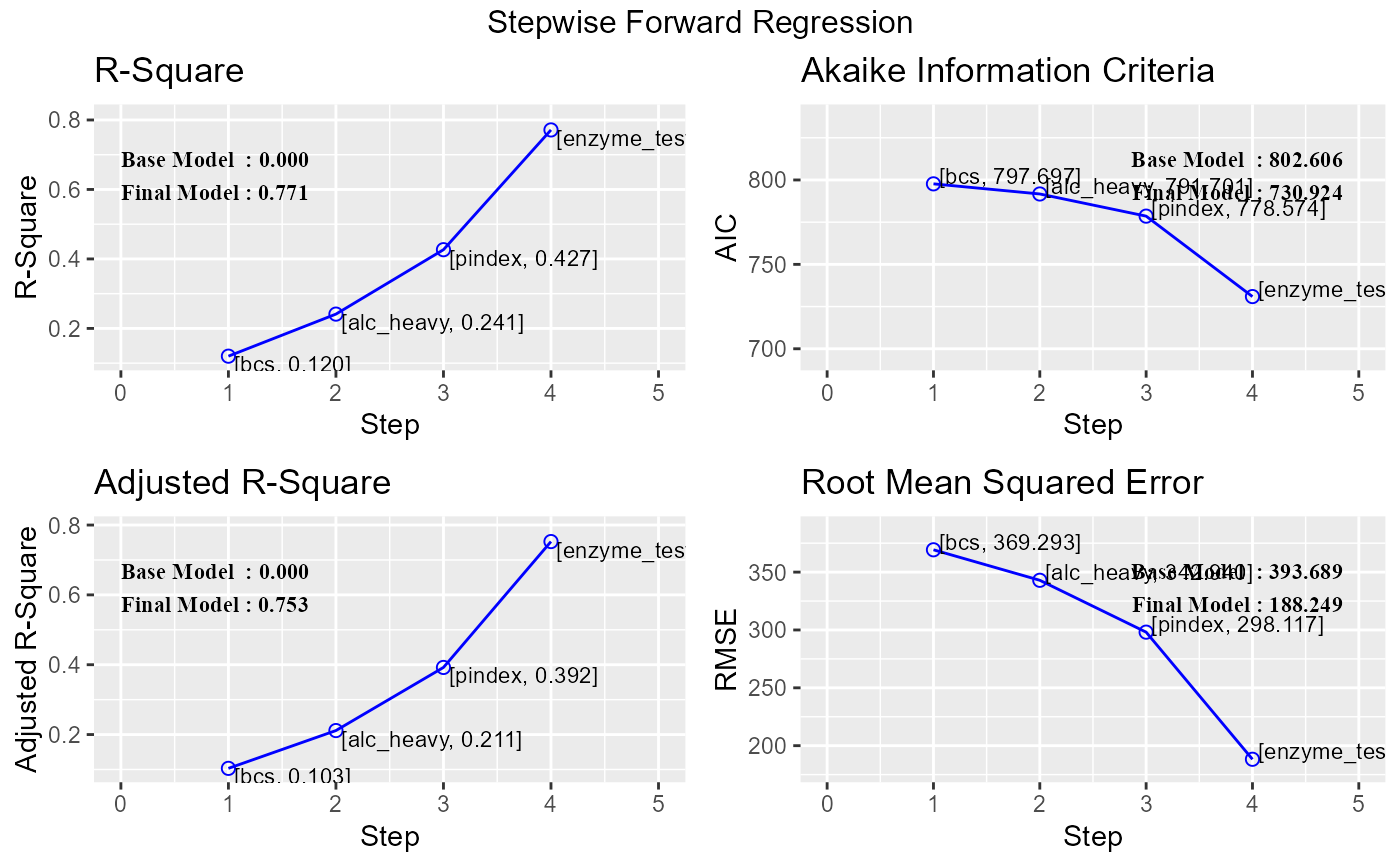
ols_step_forward_p(model, 0.1, hierarchical = TRUE)
#>
#>
#> Stepwise Summary
#> --------------------------------------------------------------------------
#> Step Variable AIC SBC SBIC R2 Adj. R2
#> --------------------------------------------------------------------------
#> 0 Base Model 802.606 806.584 646.746 0.00000 0.00000
#> 1 bcs 797.697 803.664 640.579 0.12010 0.10318
#> 2 alc_heavy 791.701 799.657 633.556 0.24119 0.21144
#> 3 pindex 778.574 788.519 620.215 0.42659 0.39218
#> 4 enzyme_test 730.924 742.858 578.678 0.77136 0.75269
#> --------------------------------------------------------------------------
#>
#> Final Model Output
#> ------------------
#>
#> Model Summary
#> -------------------------------------------------------------------
#> R 0.878 RMSE 188.249
#> R-Squared 0.771 MSE 35437.709
#> Adj. R-Squared 0.753 Coef. Var 28.147
#> Pred R-Squared 0.695 AIC 730.924
#> MAE 140.619 SBC 742.858
#> -------------------------------------------------------------------
#> RMSE: Root Mean Square Error
#> MSE: Mean Square Error
#> MAE: Mean Absolute Error
#> AIC: Akaike Information Criteria
#> SBC: Schwarz Bayesian Criteria
#>
#> ANOVA
#> -----------------------------------------------------------------------
#> Sum of
#> Squares DF Mean Square F Sig.
#> -----------------------------------------------------------------------
#> Regression 6455884.265 4 1613971.066 41.327 0.0000
#> Residual 1913636.272 49 39053.801
#> Total 8369520.537 53
#> -----------------------------------------------------------------------
#>
#> Parameter Estimates
#> ------------------------------------------------------------------------------------------------
#> model Beta Std. Error Std. Beta t Sig lower upper
#> ------------------------------------------------------------------------------------------------
#> (Intercept) -1334.424 180.589 -7.389 0.000 -1697.332 -971.516
#> bcs 81.439 17.781 0.329 4.580 0.000 45.706 117.171
#> alc_heavy 312.777 72.341 0.309 4.324 0.000 167.402 458.152
#> pindex 10.131 1.622 0.431 6.246 0.000 6.871 13.390
#> enzyme_test 11.243 1.308 0.601 8.596 0.000 8.614 13.871
#> ------------------------------------------------------------------------------------------------
#>
# plot
k <- ols_step_forward_p(model, 0.1, hierarchical = TRUE)
plot(k)
# selection metrics
k$metrics
#> step variable r2 adj_r2 aic sbc sbic mallows_cp
#> 1 1 liver_test 0.4545389 0.4440492 771.8753 777.8423 616.0089 62.511923
#> 2 2 alc_heavy 0.5667409 0.5497504 761.4394 769.3953 605.5062 41.368078
#> 3 3 enzyme_test 0.6590000 0.6385400 750.5089 760.4538 595.2974 24.337853
#> 4 4 pindex 0.7501457 0.7297495 735.7146 747.6485 582.9426 7.537284
#> 5 5 bcs 0.7809054 0.7580831 730.6204 744.5433 579.6377 3.192498
#> rmse
#> 1 290.7604
#> 2 259.1357
#> 3 229.8956
#> 4 196.7872
#> 5 184.2762
# final model
k$model
#>
#> Call:
#> lm(formula = paste(response, "~", paste(preds, collapse = " + ")),
#> data = l)
#>
#> Coefficients:
#> (Intercept) liver_test alc_heavy enzyme_test pindex bcs
#> -1178.330 58.064 317.848 9.748 8.924 59.864
#>
# include or exclude variables
# force variable to be included in selection process
ols_step_forward_p(model, include = c("age", "alc_mod"))
#>
#>
#> Stepwise Summary
#> ---------------------------------------------------------------------------
#> Step Variable AIC SBC SBIC R2 Adj. R2
#> ---------------------------------------------------------------------------
#> 0 Base Model 804.340 812.295 645.675 0.04110 0.00350
#> 1 age 803.834 809.801 646.572 0.01420 -0.00476
#> 2 alc_mod 804.340 812.295 645.675 0.04110 0.00350
#> 3 liver_test 772.922 782.867 615.246 0.48357 0.45258
#> 4 enzyme_test 763.665 775.599 606.382 0.58074 0.54652
#> 5 alc_heavy 754.332 768.255 598.224 0.66012 0.62471
#> 6 pindex 739.680 755.592 587.108 0.75031 0.71843
#> ---------------------------------------------------------------------------
#>
#> Final Model Output
#> ------------------
#>
#> Model Summary
#> -------------------------------------------------------------------
#> R 0.866 RMSE 196.724
#> R-Squared 0.750 MSE 38700.429
#> Adj. R-Squared 0.718 Coef. Var 30.034
#> Pred R-Squared 0.649 AIC 739.680
#> MAE 146.418 SBC 755.592
#> -------------------------------------------------------------------
#> RMSE: Root Mean Square Error
#> MSE: Mean Square Error
#> MAE: Mean Absolute Error
#> AIC: Akaike Information Criteria
#> SBC: Schwarz Bayesian Criteria
#>
#> ANOVA
#> -----------------------------------------------------------------------
#> Sum of
#> Squares DF Mean Square F Sig.
#> -----------------------------------------------------------------------
#> Regression 6279697.346 6 1046616.224 23.538 0.0000
#> Residual 2089823.191 47 44464.323
#> Total 8369520.537 53
#> -----------------------------------------------------------------------
#>
#> Parameter Estimates
#> -----------------------------------------------------------------------------------------------
#> model Beta Std. Error Std. Beta t Sig lower upper
#> -----------------------------------------------------------------------------------------------
#> (Intercept) -814.092 213.222 -3.818 0.000 -1243.041 -385.144
#> age 0.458 2.706 0.013 0.169 0.866 -4.985 5.902
#> alc_mod 1.088 67.941 0.001 0.016 0.987 -135.591 137.768
#> liver_test 126.675 33.832 0.341 3.744 0.000 58.613 194.737
#> enzyme_test 7.523 1.543 0.402 4.874 0.000 4.418 10.628
#> alc_heavy 361.751 87.140 0.357 4.151 0.000 186.448 537.053
#> pindex 7.862 1.908 0.334 4.120 0.000 4.023 11.700
#> -----------------------------------------------------------------------------------------------
#>
# use index of variable instead of name
ols_step_forward_p(model, include = c(5, 7))
#>
#>
#> Stepwise Summary
#> ---------------------------------------------------------------------------
#> Step Variable AIC SBC SBIC R2 Adj. R2
#> ---------------------------------------------------------------------------
#> 0 Base Model 804.340 812.295 645.675 0.04110 0.00350
#> 1 age 803.834 809.801 646.572 0.01420 -0.00476
#> 2 alc_mod 804.340 812.295 645.675 0.04110 0.00350
#> 3 liver_test 772.922 782.867 615.246 0.48357 0.45258
#> 4 enzyme_test 763.665 775.599 606.382 0.58074 0.54652
#> 5 alc_heavy 754.332 768.255 598.224 0.66012 0.62471
#> 6 pindex 739.680 755.592 587.108 0.75031 0.71843
#> ---------------------------------------------------------------------------
#>
#> Final Model Output
#> ------------------
#>
#> Model Summary
#> -------------------------------------------------------------------
#> R 0.866 RMSE 196.724
#> R-Squared 0.750 MSE 38700.429
#> Adj. R-Squared 0.718 Coef. Var 30.034
#> Pred R-Squared 0.649 AIC 739.680
#> MAE 146.418 SBC 755.592
#> -------------------------------------------------------------------
#> RMSE: Root Mean Square Error
#> MSE: Mean Square Error
#> MAE: Mean Absolute Error
#> AIC: Akaike Information Criteria
#> SBC: Schwarz Bayesian Criteria
#>
#> ANOVA
#> -----------------------------------------------------------------------
#> Sum of
#> Squares DF Mean Square F Sig.
#> -----------------------------------------------------------------------
#> Regression 6279697.346 6 1046616.224 23.538 0.0000
#> Residual 2089823.191 47 44464.323
#> Total 8369520.537 53
#> -----------------------------------------------------------------------
#>
#> Parameter Estimates
#> -----------------------------------------------------------------------------------------------
#> model Beta Std. Error Std. Beta t Sig lower upper
#> -----------------------------------------------------------------------------------------------
#> (Intercept) -814.092 213.222 -3.818 0.000 -1243.041 -385.144
#> age 0.458 2.706 0.013 0.169 0.866 -4.985 5.902
#> alc_mod 1.088 67.941 0.001 0.016 0.987 -135.591 137.768
#> liver_test 126.675 33.832 0.341 3.744 0.000 58.613 194.737
#> enzyme_test 7.523 1.543 0.402 4.874 0.000 4.418 10.628
#> alc_heavy 361.751 87.140 0.357 4.151 0.000 186.448 537.053
#> pindex 7.862 1.908 0.334 4.120 0.000 4.023 11.700
#> -----------------------------------------------------------------------------------------------
#>
# force variable to be excluded from selection process
ols_step_forward_p(model, exclude = c("pindex"))
#>
#>
#> Stepwise Summary
#> --------------------------------------------------------------------------
#> Step Variable AIC SBC SBIC R2 Adj. R2
#> --------------------------------------------------------------------------
#> 0 Base Model 802.606 806.584 646.794 0.00000 0.00000
#> 1 liver_test 771.875 777.842 616.009 0.45454 0.44405
#> 2 alc_heavy 761.439 769.395 605.506 0.56674 0.54975
#> 3 enzyme_test 750.509 760.454 595.297 0.65900 0.63854
#> 4 bcs 750.782 762.716 595.377 0.66973 0.64277
#> --------------------------------------------------------------------------
#>
#> Final Model Output
#> ------------------
#>
#> Model Summary
#> -------------------------------------------------------------------
#> R 0.818 RMSE 226.248
#> R-Squared 0.670 MSE 51188.335
#> Adj. R-Squared 0.643 Coef. Var 33.829
#> Pred R-Squared 0.567 AIC 750.782
#> MAE 171.544 SBC 762.716
#> -------------------------------------------------------------------
#> RMSE: Root Mean Square Error
#> MSE: Mean Square Error
#> MAE: Mean Absolute Error
#> AIC: Akaike Information Criteria
#> SBC: Schwarz Bayesian Criteria
#>
#> ANOVA
#> -----------------------------------------------------------------------
#> Sum of
#> Squares DF Mean Square F Sig.
#> -----------------------------------------------------------------------
#> Regression 5605350.444 4 1401337.611 24.841 0.0000
#> Residual 2764170.093 49 56411.635
#> Total 8369520.537 53
#> -----------------------------------------------------------------------
#>
#> Parameter Estimates
#> ----------------------------------------------------------------------------------------------
#> model Beta Std. Error Std. Beta t Sig lower upper
#> ----------------------------------------------------------------------------------------------
#> (Intercept) -534.711 197.967 -2.701 0.009 -932.540 -136.882
#> liver_test 149.496 43.277 0.403 3.454 0.001 62.528 236.464
#> alc_heavy 292.382 86.822 0.288 3.368 0.001 117.907 466.858
#> enzyme_test 7.431 1.929 0.397 3.851 0.000 3.554 11.309
#> bcs 34.471 27.316 0.139 1.262 0.213 -20.422 89.365
#> ----------------------------------------------------------------------------------------------
#>
# use index of variable instead of name
ols_step_forward_p(model, exclude = c(2))
#>
#>
#> Stepwise Summary
#> --------------------------------------------------------------------------
#> Step Variable AIC SBC SBIC R2 Adj. R2
#> --------------------------------------------------------------------------
#> 0 Base Model 802.606 806.584 646.794 0.00000 0.00000
#> 1 liver_test 771.875 777.842 616.009 0.45454 0.44405
#> 2 alc_heavy 761.439 769.395 605.506 0.56674 0.54975
#> 3 enzyme_test 750.509 760.454 595.297 0.65900 0.63854
#> 4 bcs 750.782 762.716 595.377 0.66973 0.64277
#> --------------------------------------------------------------------------
#>
#> Final Model Output
#> ------------------
#>
#> Model Summary
#> -------------------------------------------------------------------
#> R 0.818 RMSE 226.248
#> R-Squared 0.670 MSE 51188.335
#> Adj. R-Squared 0.643 Coef. Var 33.829
#> Pred R-Squared 0.567 AIC 750.782
#> MAE 171.544 SBC 762.716
#> -------------------------------------------------------------------
#> RMSE: Root Mean Square Error
#> MSE: Mean Square Error
#> MAE: Mean Absolute Error
#> AIC: Akaike Information Criteria
#> SBC: Schwarz Bayesian Criteria
#>
#> ANOVA
#> -----------------------------------------------------------------------
#> Sum of
#> Squares DF Mean Square F Sig.
#> -----------------------------------------------------------------------
#> Regression 5605350.444 4 1401337.611 24.841 0.0000
#> Residual 2764170.093 49 56411.635
#> Total 8369520.537 53
#> -----------------------------------------------------------------------
#>
#> Parameter Estimates
#> ----------------------------------------------------------------------------------------------
#> model Beta Std. Error Std. Beta t Sig lower upper
#> ----------------------------------------------------------------------------------------------
#> (Intercept) -534.711 197.967 -2.701 0.009 -932.540 -136.882
#> liver_test 149.496 43.277 0.403 3.454 0.001 62.528 236.464
#> alc_heavy 292.382 86.822 0.288 3.368 0.001 117.907 466.858
#> enzyme_test 7.431 1.929 0.397 3.851 0.000 3.554 11.309
#> bcs 34.471 27.316 0.139 1.262 0.213 -20.422 89.365
#> ----------------------------------------------------------------------------------------------
#>
# hierarchical selection
model <- lm(y ~ bcs + alc_heavy + pindex + enzyme_test, data = surgical)
ols_step_forward_p(model, 0.1, hierarchical = TRUE)
#>
#>
#> Stepwise Summary
#> --------------------------------------------------------------------------
#> Step Variable AIC SBC SBIC R2 Adj. R2
#> --------------------------------------------------------------------------
#> 0 Base Model 802.606 806.584 646.746 0.00000 0.00000
#> 1 bcs 797.697 803.664 640.579 0.12010 0.10318
#> 2 alc_heavy 791.701 799.657 633.556 0.24119 0.21144
#> 3 pindex 778.574 788.519 620.215 0.42659 0.39218
#> 4 enzyme_test 730.924 742.858 578.678 0.77136 0.75269
#> --------------------------------------------------------------------------
#>
#> Final Model Output
#> ------------------
#>
#> Model Summary
#> -------------------------------------------------------------------
#> R 0.878 RMSE 188.249
#> R-Squared 0.771 MSE 35437.709
#> Adj. R-Squared 0.753 Coef. Var 28.147
#> Pred R-Squared 0.695 AIC 730.924
#> MAE 140.619 SBC 742.858
#> -------------------------------------------------------------------
#> RMSE: Root Mean Square Error
#> MSE: Mean Square Error
#> MAE: Mean Absolute Error
#> AIC: Akaike Information Criteria
#> SBC: Schwarz Bayesian Criteria
#>
#> ANOVA
#> -----------------------------------------------------------------------
#> Sum of
#> Squares DF Mean Square F Sig.
#> -----------------------------------------------------------------------
#> Regression 6455884.265 4 1613971.066 41.327 0.0000
#> Residual 1913636.272 49 39053.801
#> Total 8369520.537 53
#> -----------------------------------------------------------------------
#>
#> Parameter Estimates
#> ------------------------------------------------------------------------------------------------
#> model Beta Std. Error Std. Beta t Sig lower upper
#> ------------------------------------------------------------------------------------------------
#> (Intercept) -1334.424 180.589 -7.389 0.000 -1697.332 -971.516
#> bcs 81.439 17.781 0.329 4.580 0.000 45.706 117.171
#> alc_heavy 312.777 72.341 0.309 4.324 0.000 167.402 458.152
#> pindex 10.131 1.622 0.431 6.246 0.000 6.871 13.390
#> enzyme_test 11.243 1.308 0.601 8.596 0.000 8.614 13.871
#> ------------------------------------------------------------------------------------------------
#>
# plot
k <- ols_step_forward_p(model, 0.1, hierarchical = TRUE)
plot(k)